Publications
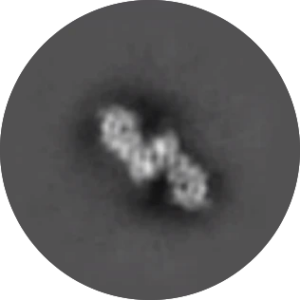 |
Structure of dimeric lipoprotein lipase reveals a pore adjacent to the active site.
Kathryn H Gunn, Saskia B Neher. Nature Communications. 2023 May |
|
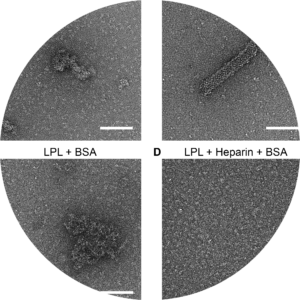 |
Combined action of albumin and heparin regulates lipoprotein lipase oligomerization, stability, and ligand interaction. | |
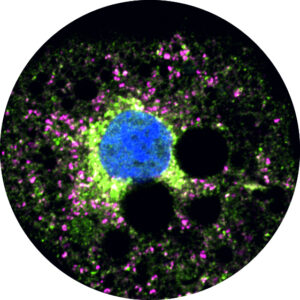 |
Characterization of lipoprotein lipase storage vesicles in 3T3-L1 adipocytes.
Roberts, B. S., Yang, C. Q., & Neher, S. B. Journal of Cell Science. 2022 |
|
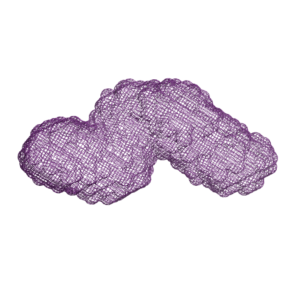 |
Comparison of angiopoietin-like protein 3 and 4 reveals structural and mechanistic similarities.
Gunn, K. H., Gutgsell, A. R., Xu, Y., Johnson, C. V., Liu, J., & Neher, S. B. Journal of Biological Chemistry. 2021 |
|
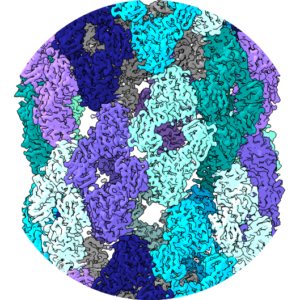 |
The structure of helical lipoprotein lipase reveals an unexpected twist in lipase storage.
Gunn K.H., Roberts, B.S., Wang F., Strauss J.D., Borgnia M.J., Egelman E.H., & Neher, S.B. Proc Natl Acad Sci. 2020 April |
|
 |
A dual apolipoprotein C-II mimetic-apolipoprotein C-III antagonist peptide lowers plasma triglycerides.
Wolska A., Lo L., Sviridov D.O., Pourmousa M., Pryor M., Ghoush S.S., Kakkar R., Davidson M, Wilson S, Pastor R.W., Goldberg I.J., Basu D., Drake S.K., Cougnoux A., Wu M.J., Neher S.B., Freeman L.A., Tang J., Amar M., Devalaraja M., & Remaley A.T. Science Translational Medicine. 2020 January |
|
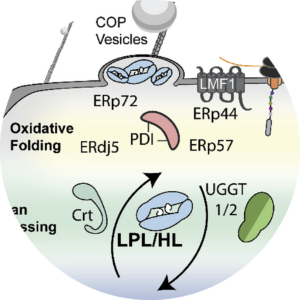 |
Endoplasmic reticulum quality control in lipoprotein metabolism. Koerner, C. M., Roberts, B. S., & Neher, S. B. Molecular and Cellular Endocrinology. 2019 |
|
 |
Protecting activity of desiccated enzymes.
Piszkiewicz, S., Gunn, K. H., Warmuth, O., Propst, A., Mehta, A., Nguyen, K. H., Kuhlman, E., Guseman, A. J., Stadmiller, S. S., Boothby, T. C., Neher, S. B., & Pielak, G. J. Protein Science. 2019 |
|
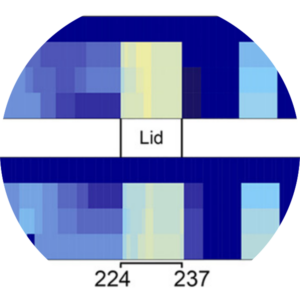 |
Mapping the sites of the lipoprotein lipase (LPL)–angiopoietin-like protein 4 (ANGPTL4) interaction provides mechanistic insight into LPL inhibition.
Gutgsell, A. R., Ghodge, S. V., Bowers, A. A., & Neher, S. B. Journal of Biological Chemistry. 2019 |
|
 |
Coexpression of novel furin-resistant LPL variants with lipase maturation factor 1 enhances LPL secretion and activity.
Wu, M. J., Wolska, A., Roberts, B. S., Pearson, E. M., Gutgsell, A. R., Remaley, A. T., & Neher, S. B. Journal of Lipid Research. 2018 |
|
 |
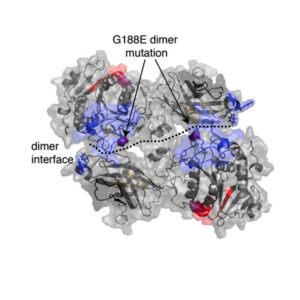
Lipase maturation factor 1 affects redox homeostasis in the endoplasmic reticulum.
Roberts, B. S., Babilonia‐Rosa, M. A., Broadwell, L. J., Wu, M. J., & Neher, S. B. The EMBO Journal. 2018 |
|
 |
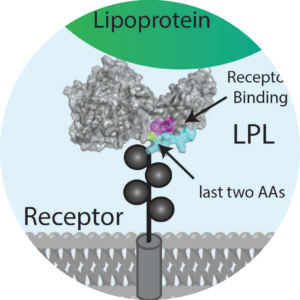
We FRET so You Don’t Have To: New Models of the Lipoprotein Lipase Dimer.
Hayne, C. K., Yumerefendi, H., Cao, L., Gauer, J. W., Lafferty, M. J., Kuhlman, B., Erie, D. A., & Neher, S. B. Biochemistry. 2018 |
|
 |
Biochemical Analysis of the Lipoprotein Lipase Truncation Variant, LPLS447X, Reveals Increased Lipoprotein Uptake.
Hayne, C. K., Lafferty, M. J., Eglinger, B. J., Kane, J. P., & Neher, S. B. Biochemistry. 2017 |
|
 |
Modulation of the Activity of Mycobacterium tuberculosis LipY by Its PE Domain. Garrett CK, Broadwell LJ, Hayne CK, Neher SB. PLoS One. 2015 August |
|
 |
Purification, cellular levels, and functional domains of Lipase Maturation Factor 1. Babilonia-Rosa, M. and Neher, SB BBRC, 2014, July |
|
 |
Signal Recognition Particle-Ribosome Binding Is Sensitive To Nascent-Chain Length.
Noriega TR, Tsai A, Elvekrog MM, Petrov A, Neher SB, Chen J, Bradshaw N, Puglisi JD, Walter P. J Biol Chem. 2014 May |
|
 |
Heat shock transcription factor σ(32) co-opts the signal recognition particle to regulate protein homeostasis in E. coli.
Lim B, Miyazaki R, Neher S, Siegele DA, Ito K, Walter P, Akiyama Y, Yura T, Gross CA. PLoS Biol. 2013 December |
|
 |
Angiopoietin-like Protein 4 Inhibition of Lipoprotein Lipase: Evidence for Reversible Complex Formation
Lafferty MJ, Bradford KC, Erie DA and Neher SB Journal of Biological Chemistry. 2013 October |
|
 |
Concerted Complex Assembly and GTPase Activation in the Chloroplast Signal Recognition Particle Biochemistry.
Nguyen TX, Chandrasekar S, Neher S, Walter P, Shan SO Biochemistry. 2011 August |
|
 |
Signal sequences activate the catalytic switch of SRP RNA
Bradshaw N*, Neher SB*, Booth DA, Walter, P. *equal contribution Science. 2009 January |
|
 |
SRP RNA controls a conformational switch regulating the SRP-SRP receptor interaction.
Neher SB*, Bradshaw N*, Floor S, Gross JD, Walter P. *equal contribution Nature Structural and Molecular Biology. 2008 September |
|
 |
Ligand-controlled proteolysis of the Escherichia coli transcriptional regulator ZntR.
Pruteanu M, Neher SB, Baker TA. J Bacteriol. 2007 April |
|
 |
Neher SB, Villen J, Oakes EC, Bakalarski CE, Sauer RT, Gygi SP, Baker TA. Mol Cell. 2006 April |
|
 |
Sculpting the proteome with AAA(+) proteases and disassembly machines.
Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Cell. 2004 October |
|
 |
Neher SB, Sauer RT, Baker TA. Proc Natl Acad Sci U S A. 2003 Nov 11 |
|
 |
Latent ClpX-recognition signals ensure LexA destruction after DNA damage.
Neher SB, Flynn JM, Sauer RT, Baker TA. Genes Dev. 2003 May |
|
 |
Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Mol Cell. 2003 March |
|
